Data Cleaning
Handling Missing Values
Sometimes, your dataset may have gaps or missing information.
Identifying Missing Values:
Start by identifying where your data has missing values. These are usually represented as blanks, "NaN" (Not-a-Number), or other placeholders.
isna()checks if each element in a DataFrame is null (missing).
- It returns a DataFrame of Boolean values (
Trueif null,Falseotherwise).
import pandas as pd
data = {'A': [1, 2, None, 4],
'B': [None, 5, 6, 7]}
df = pd.DataFrame(data)
# Check for null values in the DataFrame
null_mask = df.isna()
print(null_mask)
# Output:
A B
0 False True
1 False False
2 True False
3 False False
isna().sum() calculates the total number of missing values in each column of a DataFrame.
# Calculate the total number of null values in each column
null_counts = df.isna().sum()
print(null_counts)
# Output:
A 1
B 1
dtype: int64
In this example, 'A' has 1 missing value, and 'B' has 1 missing value.
Handling Strategies:
There are several ways to handle missing values:
1. Deletion: If missing values are less than or equal to 5% of the total values in the dataset and won't impact your analysis significantly, you can delete rows or columns with missing values.
import pandas as pd
data = {'A': [1, 2, None, 4, 7, 2, 9, 1, 7, 10, 55, 33, 7, 24, 87, 4, 7, 2, 9, 1, 7, 10, 55, 33, 7, 24, 87],
'B': [None, 5, 6, 7, 4, 1, 9, 3, 5, 3, 6, 3, 23, 56, 56, 4, 7, 2, 9, 1, 7, 10, 55, 33, 7, 24, 87]}
df = pd.DataFrame(data)
print(df)
null_values = df["A"].isna().sum()
print("null_values:", null_values)
threshold = len(df["A"]) * 0.05
print("threshold:", threshold)
# Drop rows with null values in the 'A' column
if null_values <= threshold:
df.dropna(subset = ["A"], axis = 0, inplace = True)
print(df)
# Output:
A B
0 1.0 NaN
1 2.0 5.0
2 NaN 6.0
3 4.0 7.0
4 7.0 4.0
5 2.0 1.0
6 9.0 9.0
7 1.0 3.0
8 7.0 5.0
9 10.0 3.0
10 55.0 6.0
11 33.0 3.0
12 7.0 23.0
13 24.0 56.0
14 87.0 56.0
15 4.0 4.0
16 7.0 7.0
17 2.0 2.0
18 9.0 9.0
19 1.0 1.0
20 7.0 7.0
21 10.0 10.0
22 55.0 55.0
23 33.0 33.0
24 7.0 7.0
25 24.0 24.0
26 87.0 87.0
null_values: 1
threshold: 1.35
A B
0 1.0 NaN
1 2.0 5.0
3 4.0 7.0
4 7.0 4.0
5 2.0 1.0
6 9.0 9.0
7 1.0 3.0
8 7.0 5.0
9 10.0 3.0
10 55.0 6.0
11 33.0 3.0
12 7.0 23.0
13 24.0 56.0
14 87.0 56.0
15 4.0 4.0
16 7.0 7.0
17 2.0 2.0
18 9.0 9.0
19 1.0 1.0
20 7.0 7.0
21 10.0 10.0
22 55.0 55.0
23 33.0 33.0
24 7.0 7.0
25 24.0 24.0
26 87.0 87.0
2. Imputation: Fill in missing values with appropriate replacements if they are more than 5% of the total values in the dataset. This can be done using the mean, median, mode, or even more complex imputation methods based on the nature of your data.
import pandas as pd
data = {'A': [1, 2, None, 9, 1, 7, 10, 55, 33, 7, 24, 87, 4, 7, 2, 9, 1, 7, 10],
'B': [None, 4, 1, 9, 3, 5, 3, 6, 3, 23, 56, 56, 4, 7, 2, 9, 1, 7, 10]}
df = pd.DataFrame(data)
print(df)
null_values = df["A"].isna().sum()
print("null_values:", null_values)
threshold = len(df["A"]) * 0.05
print("threshold:", threshold)
# Fill null values with Mean in the 'A' column
if null_values > threshold:
df["A"].fillna(df["A"].mean(), inplace = True)
print(df)
# Output:
A B
0 1.0 NaN
1 2.0 4.0
2 NaN 1.0
3 9.0 9.0
4 1.0 3.0
5 7.0 5.0
6 10.0 3.0
7 55.0 6.0
8 33.0 3.0
9 7.0 23.0
10 24.0 56.0
11 87.0 56.0
12 4.0 4.0
13 7.0 7.0
14 2.0 2.0
15 9.0 9.0
16 1.0 1.0
17 7.0 7.0
18 10.0 10.0
null_values: 1
threshold: 0.9500000000000001
A B
0 1.000 NaN
1 2.000 4.0
2 15.333 1.0
3 9.000 9.0
4 1.000 3.0
5 7.000 5.0
6 10.000 3.0
7 55.000 6.0
8 33.000 3.0
9 7.000 23.0
10 24.000 56.0
11 87.000 56.0
12 4.000 4.0
13 7.000 7.0
14 2.000 2.0
15 9.000 9.0
16 1.000 1.0
17 7.000 7.0
18 10.000 10.0
Imputing by Sub-group:
Let’s we have a dataset with the following columns:
| Experience | Experience Level e.g., “Mid”, “Senior” |
|---|---|
| Salary | Salary in USD dollars |
salaries_dict = salaries.groupby("Experience")["Salary_USD"].median().to_dict()
print(salaries_dict)
# {'Entry': 55380.0, 'Executive': 135439.0, 'Mid': 74173.5, 'Senior': 128903.0}
salaries["Salary_USD"] = salaries["Salary_USD"].fillna(salaries["Experience"].map(salaries_dict))
# The .map(salaries_dict) part creates a new Series where each experience in the "Experience" column is replaced with the corresponding median salary from salaries_dict.
# Essentially, if a row in the "Salary_USD" column is missing, it will be filled with the median salary of the corresponding experience.
Removing Duplicate Values
Duplicates in your data can skew your analysis.
Identifying Duplicates:
Detect duplicate records by comparing rows to see if they have identical values across all or specific columns.
Handling Strategies:
Dropping Duplicates: Remove duplicate records, keeping only the first occurrence.
import pandas as pd
data = {'Name': ["Ali", "Bilal", "Usma", "Bilal", "Sana", "Alishba"],
'Age': [15, 18, 21, 18, 20, 19]}
df = pd.DataFrame(data)
print(df)
Name Age
0 Ali 15
1 Bilal 18
2 Usma 21
3 Bilal 18
4 Sana 20
5 Alishba 19
df = df.drop_duplicates(subset = "Name", keep='first')
print(df)
Name Age
0 Ali 15
1 Bilal 18
2 Usma 21
4 Sana 20
5 Alishba 19
Identifying Outliers
Outliers are data points that are significantly different from the rest of the data.
Identifying Outliers:
Visualizations like box plots, scatter plots, or statistical methods can help identify them. We can also identify outliers using mathematical methods like:
1. Z-Score Method: Any data point with a Z-Score greater than 3 or less than -3 is considered an outlier.
import pandas as pd
# Sample data
data = {'Value': [10, 12, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 100]}
# Convert data to a pandas DataFrame
df = pd.DataFrame(data)
# Function to calculate Z-Score
def calculate_z_scores(df):
mean = df['Value'].mean()
std_dev = df['Value'].std()
df['Z-Score'] = (df['Value'] - mean) / std_dev
return df
# Function to identify outliers using Z-Score method
def z_score_outliers(df):
df = calculate_z_scores(df)
outliers = df[(df['Z-Score'] > 3) | (df['Z-Score'] < -3)]
return outliers
# Identify outliers
z_score_outliers_df = z_score_outliers(df.copy())
print("Original Data:\n", df)
print("Z-Score Outliers:", z_score_outliers_df)
Original Data:
Value
0 10
1 12
2 14
3 15
4 16
5 17
6 18
7 19
8 20
9 21
10 22
11 23
12 24
13 100
Z-Score Outliers:
Value Z-Score
13 100 3.415
- Lower Bound: Q1 - 1.5 * IQR
- Upper Bound: Q3 + 1.5 * IQR
- Any data point below lower bound or above upper bound would be considered an outlier.
import pandas as pd
# Sample data
data = {'Value': [10, 12, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 100]}
# Convert data to a pandas DataFrame
df = pd.DataFrame(data)
# Function to identify outliers using IQR method
def iqr_outliers(df):
Q1 = df['Value'].quantile(0.25)
Q3 = df['Value'].quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
outliers = df[(df['Value'] < lower_bound) | (df['Value'] > upper_bound)]
return outliers
# Identify outliers
iqr_outliers_df = iqr_outliers(df.copy())
print("Original Data:\n", df)
print("\nIQR Outliers:\n", iqr_outliers_df)
Original Data:
Value
0 10
1 12
2 14
3 15
4 16
5 17
6 18
7 19
8 20
9 21
10 22
11 23
12 24
13 100
IQR Outliers:
Value
13 100
Handling Strategies:
1. Remove Outliers: In some cases, outliers may be data entry errors or anomalies. Removing them might be appropriate.
import pandas as pd
# Sample data
data = {'Value': [10, 12, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 100]}
# Convert data to a pandas DataFrame
df = pd.DataFrame(data)
# Function to calculate Z-Score
def calculate_z_scores(df):
mean = df['Value'].mean()
std_dev = df['Value'].std()
df['Z-Score'] = (df['Value'] - mean) / std_dev
return df
# Function to identify outliers using Z-Score method
def z_score_outliers(df):
df = calculate_z_scores(df)
outliers = df[(df['Z-Score'] > 3) | (df['Z-Score'] < -3)]
return outliers
# Function to identify outliers using IQR method
def iqr_outliers(df):
Q1 = df['Value'].quantile(0.25)
Q3 = df['Value'].quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
outliers = df[(df['Value'] < lower_bound) | (df['Value'] > upper_bound)]
return outliers
# Function to remove outliers
def remove_outliers(df, outliers):
cleaned_df = df[~df.isin(outliers)].dropna()
return cleaned_df
# Function to smooth outliers
def smooth_outliers(df, outliers):
median_value = df['Value'].median()
df.loc[df.index.isin(outliers.index), 'Value'] = median_value
return df
# Identify outliers
z_score_outliers_df = z_score_outliers(df.copy())
iqr_outliers_df = iqr_outliers(df.copy())
# Handle outliers by removing them
cleaned_df = remove_outliers(df.copy(), iqr_outliers_df)
# Handle outliers by smoothing them
smoothed_df = smooth_outliers(df.copy(), iqr_outliers_df)
print("Original Data:\n", df)
print("\nZ-Score Outliers:\n", z_score_outliers_df)
print("\nIQR Outliers:\n", iqr_outliers_df)
print("\nData after Removing Outliers:\n", cleaned_df)
print("\nData after Smoothing Outliers:\n", smoothed_df)
2. Smooth Data: Change outliers with appropriate replacements. This can be done using the median or mode etc.
import numpy as np
import pandas as pd
# Sample data
data = {'Value': [10, 12, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 100]}
# Convert data to a pandas DataFrame
df = pd.DataFrame(data)
# Function to identify and smooth outliers using IQR method
def smooth_outliers(df):
Q1 = df['Value'].quantile(0.25)
Q3 = df['Value'].quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
median_value = df['Value'].median()
df['Value'] = np.where((df['Value'] < lower_bound) | (df['Value'] > upper_bound), median_value, df['Value'])
return df
# Handle outliers by smoothing them
smoothed_df = smooth_outliers(df.copy())
print("Original Data:\n", df)
print("\nData after Smoothing Outliers:\n", smoothed_df)
Original Data:
Value
0 10
1 12
2 14
3 15
4 16
5 17
6 18
7 19
8 20
9 21
10 22
11 23
12 24
13 100
Data after Smoothing Outliers:
Value
0 10.0
1 12.0
2 14.0
3 15.0
4 16.0
5 17.0
6 18.0
7 19.0
8 20.0
9 21.0
10 22.0
11 23.0
12 24.0
13 18.5
Correcting Inconsistent Data
Identifying Inconsistencies:
Look for inconsistencies in data, such as variations in formatting, spelling errors, or units of measurement.
Handling Strategies:
Correcting inconsistencies involves:
- Standardization: Ensure consistent formatting for text data (e.g., capitalization) and dates.
- Data Validation: Validate data against predefined rules or patterns to catch inconsistencies.
- Conversion: Convert units of measurement to a consistent format.
- Imputing Correct Data: Replace incorrect data with the correct values when possible.
Data Munging
Data munging is the process of transforming, and organizing raw data into a format that is suitable for analysis or further processing.
Data Munging Tasks:
- Renaming Variables
- Data Type Conversion
- Encoding, Decoding, Recoding data
- Data Normalization
Renaming Variable
Renaming variables in data munging involves changing the names of columns or variables in a dataset. This is done for several reasons, including making the variable names more meaningful, clear, or consistent with your analysis goals. Here are some common scenarios where you might need to rename variables:
- Clarity: Variable names may be abbreviated in the original dataset, making it difficult to understand their meaning. Renaming them to more descriptive names can improve clarity.
- Consistency: Ensuring that variable names follow a consistent naming convention across your dataset or with other datasets can make it easier to work with and merge data.
- Standardization: In some cases, you might need to standardize variable names to match industry or organizational standards.
Here's an example of how you can rename variables using Python's pandas library:
import pandas as pd
# Create a sample DataFrame
data = {'Old_Variable_Name1': [1, 2, 3],
'Old_Variable_Name2': [4, 5, 6]}
df = pd.DataFrame(data)
# To get Columns Name
print(df.columns)
# Rename variables
df.rename(columns={'Old_Variable_Name1': 'New_Variable_Name1',
'Old_Variable_Name2': 'New_Variable_Name2'}, inplace = True)
# Now, the DataFrame has variables with new names
Data Type Conversion
Data type conversion, also known as data type casting, is the process of changing the type of data in a column to ensure it's compatible with your analysis or processing needs. This is a crucial data munging task because datasets often contain a mix of different data types, and converting them to the right type is essential for accurate analysis. Here are some common scenarios where data type conversion is necessary:
- String to Numeric: Converting string values (text) to numeric types (integers or floating-point numbers) is common when working with numerical data. For example, you might need to convert "123" to the integer 123.
- Numeric to String: Sometimes, you may need to convert numeric values to strings, especially when you want to include them in text output or concatenate them with other strings.
- Date and Time Formats: Converting date and time data to a consistent format (e.g., from text to datetime objects) allows for date-based analysis and calculations.
- Categorical to Numerical: Converting categorical data (e.g., "low," "medium," "high") to numerical values (e.g., 0, 1, 2) for use in machine learning algorithms.
- Boolean Conversion: Converting binary values (e.g., "yes" or "no") to boolean values (True or False).
Here's an example in Python using pandas to convert a data type:
import pandas as pd
# Convert data type from string to integer
data = {'Column1': ['123', '456', '789']}
df = pd.DataFrame(data)
print(df.dtypes)
# Column1 object
# dtype: object
df['Column1'] = df['Column1'].astype(int)
print(df.dtypes)
# Column1 int64
# dtype: object
# Convert data type from string to datetime
data = {'Column1': ['2024-06-26', '2023-02-02', '2020-01-22']}
df = pd.DataFrame(data)
print(df.dtypes)
# Column1 object
# dtype: object
df['Column1'] = pd.to_datetime(df['Column1'])
print(df.dtypes)
# Column1 datetime64[ns]
# dtype: object
Encoding, Decoding, and Recoding Data
Encoding, decoding, and recoding data are data munging tasks that deal with converting and transforming categorical variables, text, or other non-numeric data into a format that can be used for analysis or modeling.
Encoding Data:
Encoding involves converting categorical or textual data into numerical representations. This is essential when working with machine learning algorithms that require numerical inputs. For example, assign a unique integer to each category. It is suitable for ordinal categorical data (categories with a specific order).
import pandas as pd
# Sample data
data = pd.DataFrame({'Category': ['Low', 'Medium', 'High', 'Medium', 'Low']})
# Create a mapping of categories to integers
category_mapping = {category: idx for idx, category in enumerate(data['Category'].unique())}
# Encode the data
data['Encoded'] = data['Category'].map(category_mapping)
print(data)
# Output:
# Category Encoded
# 0 Low 0
# 1 Medium 1
# 2 High 2
# 3 Medium 1
# 4 Low 0
Decoding Data:
Decoding is the reverse process of encoding. It converts numerical representations back into their original categorical or textual form. This is often done for interpretability or reporting purposes.
# Decode the data
data['Decoded'] = data['Encoded'].map({v: k for k, v in category_mapping.items()})
print(data)
# Output:
# Category Encoded Decoded
# 0 Low 0 Low
# 1 Medium 1 Medium
# 2 High 2 High
# 3 Medium 1 Medium
# 4 Low 0 Low
Recoding Data:
Recoding refers to changing the values of a variable to make them more meaningful, manageable, or suitable for analysis. This can involve combining categories, creating new categories, or mapping specific values to others.
# Sample data
data = pd.DataFrame({'Age': [10, 25, 67, 45, 33]})
# Define bins and labels for age groups
bins = [0, 18, 35, 60, 100]
labels = ['Child', 'Young Adult', 'Adult', 'Senior']
# Recoding age into age groups
data['Age Group'] = pd.cut(data['Age'], bins=bins, labels=labels)
print(data)
# Output:
# Age Age Group
# 0 10 Child
# 1 25 Young Adult
# 2 67 Senior
# 3 45 Adult
# 4 33 Young Adult
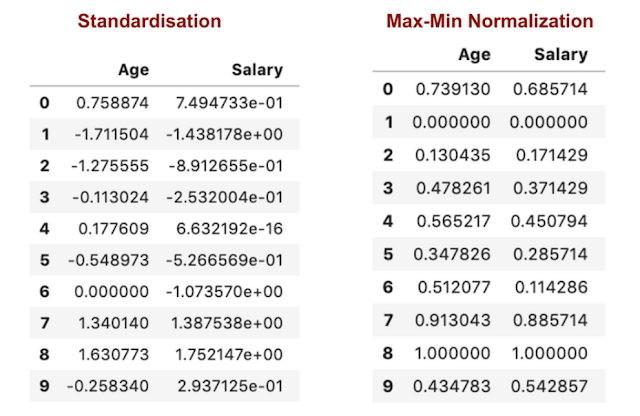
Data Normalization
Example:
1. Min-Max Normalization:
Min-Max scaling is a common normalization technique that re-scales features to a distribution value between 0 and 1. The minimum value transforms into 0, and the maximum value transforms into 1. This implies the data is more concentrated around the mean with Max-Min Normalization. If there are outliers in your features, normalizing data scales most of it to a small interval, providing uniform scales but not handling outliers well. Standardization is more robust to outliers, making it preferable in many cases.
from sklearn.preprocessing import MinMaxScaler
import numpy as np
# Example data
data = np.array([[100], [200], [300], [100], [400]])
# Initialize MinMaxScaler
scaler = MinMaxScaler()
# Perform Min-Max Normalization
normalized_data = scaler.fit_transform(data)
print("Min-Max Normalized Data:")
print(normalized_data)
2. Standardization (Z-Score Normalization):
Rescales features to ensure mean and standard deviation are 0 and 1, respectively. The range is from -3 to +3.
from sklearn.preprocessing import StandardScaler
import numpy as np
# Example data
data = np.array([[100], [200], [300], [100], [400]])
# Initialize StandardScaler
scaler = StandardScaler()
# Perform Standardization
standardized_data = scaler.fit_transform(data)
print("Standardized Data (Z-Score):")
print(standardized_data)